Official Journals By StatPerson Publication
|
Table of Content Volume 8 Issue 2 - November 2018
Amoxicillin clavulanic acid resistance in pus samples sent for culture sensitivity - The scenario in a tertiary care hospital, Ballari
Murgesh J V1, Vishwanath M2*, Prashanth S P3
1Associate Professor, 2Assistant Professor, 3Post Graduate, Department of Pharmacology, Vijayanagar Institute of Medical Sciences, Ballari, Karnataka, INDIA. Email: vishwa.m100@gmail.com
Abstract Background: Amoxicillin Clavulanic Acid (AMC) is a broad-spectrum antibacterial being used over 20 years. Emergence of resistant bacteria has complicated the empirical therapy of infections. Resistance to Amoxicillin is a matter of concern. Clavulanic acid, a β-lactamase inhibitor, is used to overcome resistance. But appearance of resistance to even this combination is a matter of even more concern. Hence an attempt is made to find out the proportion of resistance to AMC in a tertiary care setup. Aim: To evaluate the pattern of resistance to Amoxicillin Clavulanic acid (AMC) among organisms isolated from pus samples sent for culture sensitivity. Material and Methods: This record based descriptive study, taken up to evaluate the resistance pattern of culture isolates of pus samples from various departments. Pus culture and sensitivity reports obtained by Kirby Bauer Disc diffusion method of pus samples during a period of 6 months was collected from Microbiology Department. Data obtained were tabulated and analysed using descriptive statistical methods. Results: A total of 245 reports were analysed. Most common organism isolated (I) and its resistance (R) was Staphylococcus (I:46.53%; R:61.4%), followed by E.coli (I:19.59%; R:87.5%), Pseudomonas (I:14.29%; R:62.85%), Klebsiella (I:11.43%; R:85.71%), Citrobacter (I:3.27%; R:100%), Streptococci (I:2.04%; R:0%), Proteus (I:1.63%; R:75%), Enterobacter (I:0.82%; R:50%) and Acetobacter (I:0.4%; R:0%). 69.4% of sample culture isolates were found to be resistant to AMC. Conclusion: This study reports the most commonly encountered organism in pus samples to be Staphylococcus, followed by E.coli, Pseudomonas and Klebsiella. Nearly 70% of organisms causing purulent infections were resistant to AMC. Of the isolates, statistically significant resistance was shown by E.coli followed by Klebsiella and Staphylococcus. Key Word: Antibiotic, Amoxicillin clavulanic acid, resistance, culture sensitivity
INTRODUCTION Bacterial infections are one of the growing concerns globally, especially so when they are resistant to commonly used multiple antibiotics. Pyogenic infections can either be mono or poly microbial, aerobic and anaerobic or a mixture of both involving an average of 5-6 organisms on culture. 1 Antibiotics are undoubtedly a blessing to human civilization that has saved millions of lives fighting against microbes or infections. 2 The first β -lactams antibiotics were introduced for use in the 1950s (Penicillin G and V), but they had substantial inconveniences, like a limited range of activity, a short half-life, and the administration route had to be parenteral. 3 To overcome these, amoxicillin was developed and marketed in 1972, which was a moderate spectrum β -lactam active against wide range of Gram-positive and a limited range of Gram-negative organisms. It still remains the most commonly utilised drug in the class due to its better oral availability.4 During the past decades many bacteria developed the ability to resist β -Lactam antibiotics, with β -Lactamase production being the most common mechanism of resistance. β -Lactamase produced by several Gram-positive organisms (including Staphylococci spp.), Gram-negative organisms (Escherichia coli, Neisseria gonorrhoeae, Haemophilus influenzae, Moraxella catarrhalis, Pseudomonas and Klebsiella spp.), and anaerobic organisms (Bacteroides spp.) can cleave the β -lactam ring and render the antibiotic inactive.5 To overcome the problem of β -Lactamase production, research was directed towards identifying compounds which can inhibit β -lactamase. In 1972, a compound called Clavulanic Acid was identified which was structurally related to penicillins and prevented inactivation of β -Lactam antibiotics when given together.6 The combination of amoxycillin and clavulanic acid (AMC) was made available to market in 1981, which was the only penicillin and β -lactamase inhibitor available in oral formulation.7 But the battle between the microbes and the antibiotics did not end there. Organisms developed mechanisms to over the use of β -Lactamase inhibitors too. The practice of antibiotic misuse and prophylactic use of antibiotics in humans as well as agricultural industries has increased exposure of antibiotics to microbes facilitating even rapid development of resistance.8 Antimicrobial resistance in pathogens causing important communicable diseases has become a matter of great public health concern globally. The problem of resistance has emerged especially more so in developing countries like India where there is widespread availability of practically all the antimicrobials over the counter.9 Resistance to Amoxicillin is itself a matter of concern. Clavulanic acid, a β-lactamase inhibitor, is used to overcome resistance. But appearance of resistance to even this combination of drugs is a matter of even more concern. The type of organism causing the infection and their sensitivity and resistance pattern vary from place to place. Therefore, appropriate selection of an effective antibiotic requires knowledge of the potential microbes and its resistance and sensitivity patterns.10 Rapid detection and reporting of resistant organism, monitoring and providing feedback regarding antibiotic resistance and other measures are the important strategies to control antibiotic resistance in hospitals.11 Hence this study was taken up to evaluate the pattern of resistance to AMC among organisms isolated from pus samples sent for culture sensitivity in a tertiary care hospital.
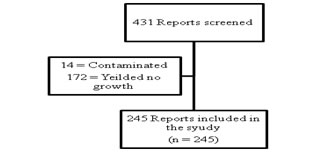
MATERIAL AND METHODS Study design: This is a record based descriptive study, taken up in the department of Pharmacology, to evaluate the resistance pattern of culture isolates of pus samples from various departments sent to the department of microbiology. Inclusion criteria: All aerobic culture and sensitivity reports obtained by Kirby Bauer Disc Diffusion method of pus samples sent to department of microbiology during the period of 6 months from February to July 2016 were included in the study. Exclusion criteria: All samples with no isolates or those reported as contaminated were excluded from the study. Methodology: After obtaining clearance from the Institutional Ethics committee, all the reports for aerobic pus culture and sensitivity obtained by Kirby Bauer Disc diffusion method from various departments were collected over a period of 6 months starting from February to July 2016, from the central laboratory at our institution. All the reports that did not have any isolates and all the reports reported as contaminated were excluded from the study (Figure 1). Statistical analysis: The data obtained were tabulated on an excel sheet and analysed using descriptive statistical methods. Figure 1: Data Flow chart. RESULTS A total of 431 reports were obtained over a period of six months from February to July 2016. Of the 431 reports only 245 samples yielded isolates and satisfied all inclusion criteria. The data obtained from these 245 reports were tabulated and analysed using descriptive statistical methods (n = 245). 56 isolates were present in the month of June followed by 50 isolates in February and 42 in the month of April. Least number of isolates was seen in the month of May accounting for 27 isolates. The month wise number of reports obtained is shown in Table 1. Table 1: Month wise distribution of results
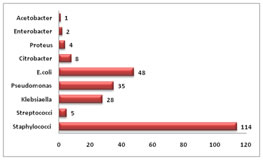
The organism that was most commonly isolated was staphylococcus114 followed by E. coli48, Pseudomonas35 and Klebsiella28.The other organisms that were isolated were Citrobacter, Proteus, Enterobacter, Acetobacter and streptococcus (Figure 2). Figure 2: Commonly isolated organisms
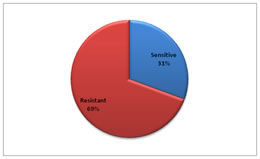
Of the 245 reports included in the study, 75 (31%) were reported sensitive to AMC whereas 170 (69%) were reported resistant (Figure 3). Figure 3: Overall resistance of isolates
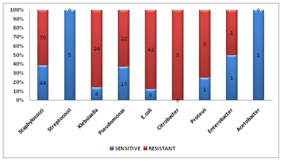
Of all the isolates, Citrobacter showed the highest rate of resistance to AMC (100%), followed by E.coli (87.5%), Klebsiella (85.71%), Proteus (75%), Pseudomonas (62.86%), Staphylococci (61.4%) and Enterobacter (50%). All the Streptococci that were isolated were sensitive to AMC. However, statistically significant resistance was identified highest in E. coli (87.5%, p=0.002), followed by Klebsiella (85.71%, p=0.046) and Staphylococci (61.4%, p=0.001) among the isolated organisms (Table 2) (Figure 4). Table 2: Percentage resistance of different organisms isolated
Figure 4: Percentage resistance of individual organisms isolated
None of the streptococci and acetobacter species that were isolated was found to be resistant to AMC. All of the citrobacter species isolated showed resistance to AMC however it was not statistically significant (p=0.056). Staphylococci which was the most commonly isolated organism had a resistance of 61.4% (p=0.001).
DISCUSSION The results of our study correlate to findings of different authors from across the country. In a study by Bansal V et al, at Post Graduate Institute of Medical Education and Research, Chandigarh, India, a significant increase was noticed in the prescription of antimicrobials in 2005 as compared with 1995 (570 vs 482, P<0.001). They noticed an increase in the use of Amoxicillin-Clavulanic acid, Metronidazole, Amikacin, and third and fourth-generation cephalosporins. Penicillin and cephalosporin remained on top of the list. They found that the combination is routinely indicated for empirical therapy of serious infections in both immunocompromised and immunecompetent individuals and has also found place in WHO Essential Medicine List (EML).12 This indicates that there is an increasing trend in prescribing higher antibiotics compared to earlier days. The findings of our study correlates with the findings of the study conducted by Banu A et al, where 100 pus samples were collected from patients with chronic diabetic foot ulcers of which 82 isolates were obtained. The most common isolated organism was Staphylococcus aureus followed by E.coli, which correlates with the findings of our study. Eighty of the isolates were multi drug resistant with majority being gram negative. They reported a resistance of 56.1% to Amoxicillin Clavulanic acid which is much lower than what was observed in our study. 13 Another study done by Jamatia A et al, a retrospective analysis of 359 consecutive pus specimens were done to evaluate the antibiotic susceptibility done by modified Kirby-Bauer methods (n=176). They reported Staphylococcus aureus to be the most common organism isolated which correlates with our study, followed by Pseudomonas spp., Klebsiella spp. and Escherichia coli. They reported a 100% resistance to amoxicillin clavulanic acid which is much higher than that present in organisms in our locality.14 All the evidences indicate the rising levels of resistance to amoxicillin clavulanic acid calling for a much judicious use of the antibiotic. Though this was a simple study, it throws light on the raising concern of antibiotic resistance being developed to a drug used to overcome resistant mechanism. This compels us to show more attention in using correct antibiotic policies as well as to increase the research on newer and more effective antibiotics. However, in our study as only the pus cultures sent for analysis to the department of Microbiology were analysed, many factors like the diabetic status of the patients, prior antibiotic use and other relevant factors that may influence resistance pattern are not accounted for. Also, significantly different susceptibility results are obtained with change in the antibiotic disc, laboratories tested, and locality.15 Hence a more clinical correlation of the results obtained is necessary for appropriate judgement. Antibiotic drug resistance is not a new phenomenon; however the current magnitude of the problem and the speed with which the new resistant phenotypes have emerged elevates the public health importance of this issue. The societal and financial costs of treating antimicrobial resistant infections place a significant human and economic burden on society as individuals infected with drug resistant organisms are more likely to remain in hospital for a longer period of time and have poor prognosis. In addition, the scarcity of new antimicrobial agents and the lack of new agents in the drug development pipeline limit treatment options, particularly for patients with infections caused by multi-drug resistant (MDR) organisms, which occur mainly in health care settings. For drug industry, it represents diminished marketability of current products. 16 As a result, it has become very important to take quick and effective steps against the emergence of antimicrobial resistance.
CONCLUSION This study reports the most commonly encountered organism in pus samples to be Staphylococcus, followed by E.coli, Pseudomonas and Klebsiella. Nearly 70% of the organisms causing purulent infections were resistant to AMC, which calls for a judicious use this broad spectrum antibiotic. Of the isolates, statistically significant resistance was shown by E.coli followed by Klebsiella and Staphylococcus. However a clinical correlation of the results needs to be considered in choice and use of antibiotics. The development of antibiotic resistance can be prevented by rational and judicious use of antibiotics. It has become inevitable for formulation of antibiotic policy for rational drug use, regular monitoring of progress in implementation of policies and guidelines related to antibiotic resistance, which could help in the modern day scenario of emerging resistance. Application of issues of “Antibiotic stewardship” effectively can limit the increasing antibiotic resistance as: 1) Indiscriminate use of antibiotics to be avoided by ensuring their indication, dose and duration of treatment. 2) Restrict the use of combination of antimicrobials to appropriate circumstances. 3) Restrict the drug use, e.g. limit the use of newest member of a group of antibiotics as long as the current drugs are effective. 4) Monitoring of resistance pattern constantly in a hospital or community. 5) Change the empirical drug therapy when needed. 6) Practices of infection control in hospitals to be made compulsory.
REFERENCES
|
|
 Home
Home